This is Blog #3 in a 3 Part Blog Series On HL7 and Healthcare Data Interoperability
In the past 2 blog posts (Click here to access Blog #1 and Blog #2) of this series, we’ve shown you how to use Smolder to parse your hl7v2 data and build a Medallion architecture within Databricks.
Let’s look at the bigger picture though. As a healthcare organization, it’s likely you have many different data sources beyond hl7v2 data. These data sources might be based on standards like FHIR or even OMOP.
This brings up some new questions: How do I take advantage of all of this data and perform quick analytics when it’s being collected in all of these different formats? How can I easily move between these different data formats? How can I integrate all of these different standards in the same pipeline?
These questions lead us to the overarching idea of healthcare interoperability: the ability to share and use data from various health standards securely and quickly.
Designing an Interoperability Tool
Developing a well-designed interoperability tool is easier said than done.
In our hypothetical architecture, we’ll start off with the assumption that each health data source is represented as its own data model. For instance, we would have a data model for FHIR, a separate data model for OMOP, and so on. In order to move from one data model to another we need to create mappings between these different data models. Let’s start off simple and look at two data models: hl7v2 and FHIR.
In the diagram below we see that we need to create two sets of mappings: 1) to go from hl7v2 to FHIR and 2) to go from FHIR to hl7v2.

OMOP
Now what if we add OMOP as a third data model into the mix. We see that we now need to implement triple the number of mappings in our architecture. This highlights a key challenge for interoperability which is that these data models have a many-to-many relationship. Implementing and maintaining all of the mappings becomes unrealistic as the number of data models grows.
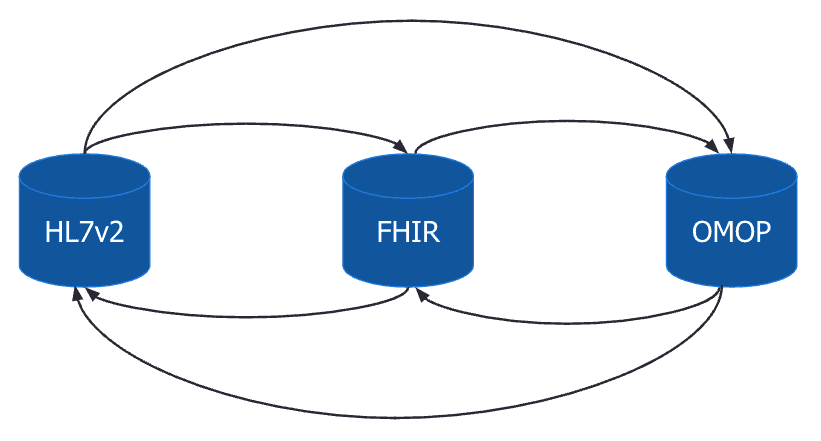
One way to overcome this issue would be through the use of an intermediate data model. Let’s continue with the previous example but set the OMOP data model as an intermediate data model.
The diagram below illustrates the new architecture. For each additional data model we only need to create a mapping between the new data model and the intermediate data model. This significantly reduces the complexity, especially as the number of data models grows, since we no longer have to map each additional data model to all existing data models.

There’s a lot more that goes into creating a well-designed healthcare interoperability tool than what we’ve discussed thus far in this blog post. So much so, that it could be the topic of its own multi-part blog post series. To continue the discussion, join us at the Databricks AI Summit where we dive deeper into interoperability, its importance, and the exciting solutions we’ve been developing to start answering the questions outlined in this post.
Lovelytics is a preferred partner of Databricks and has helped many clients install and configure their Databricks instances. To learn more please visit us at www.lovelytics.com/partners/databricks or connect with us via email at [email protected]
Healthcare Interoperability is a focus for Lovelytics and Databricks. Please join us at the Databricks AI Summit on June 27-30 2022 in San Francisco to see Healthcare Interoperability in action. Click here to register to attend.